You must login before you can run this tool.
Multicellular modeling of mRNA vaccine-loaded lipid nanoparticles for cancer immunotherapy
Simulation mRNA vaccine-loaded lipid nanoparticles (LNPs) for cancer immunotherapy with PhysiCell
Category
Published on
Abstract
LNPs-based mRNA vaccine for cancer immunotherapy
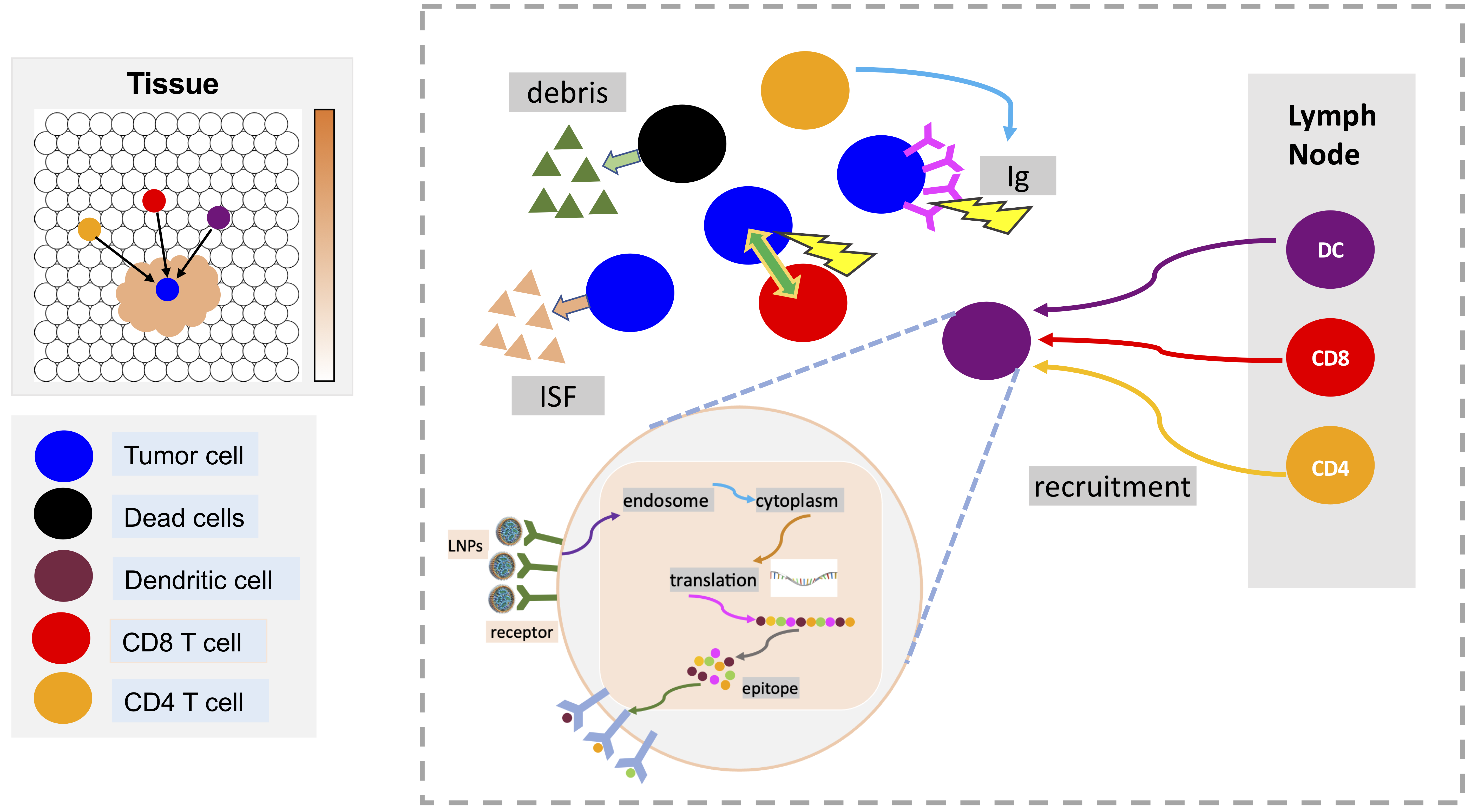
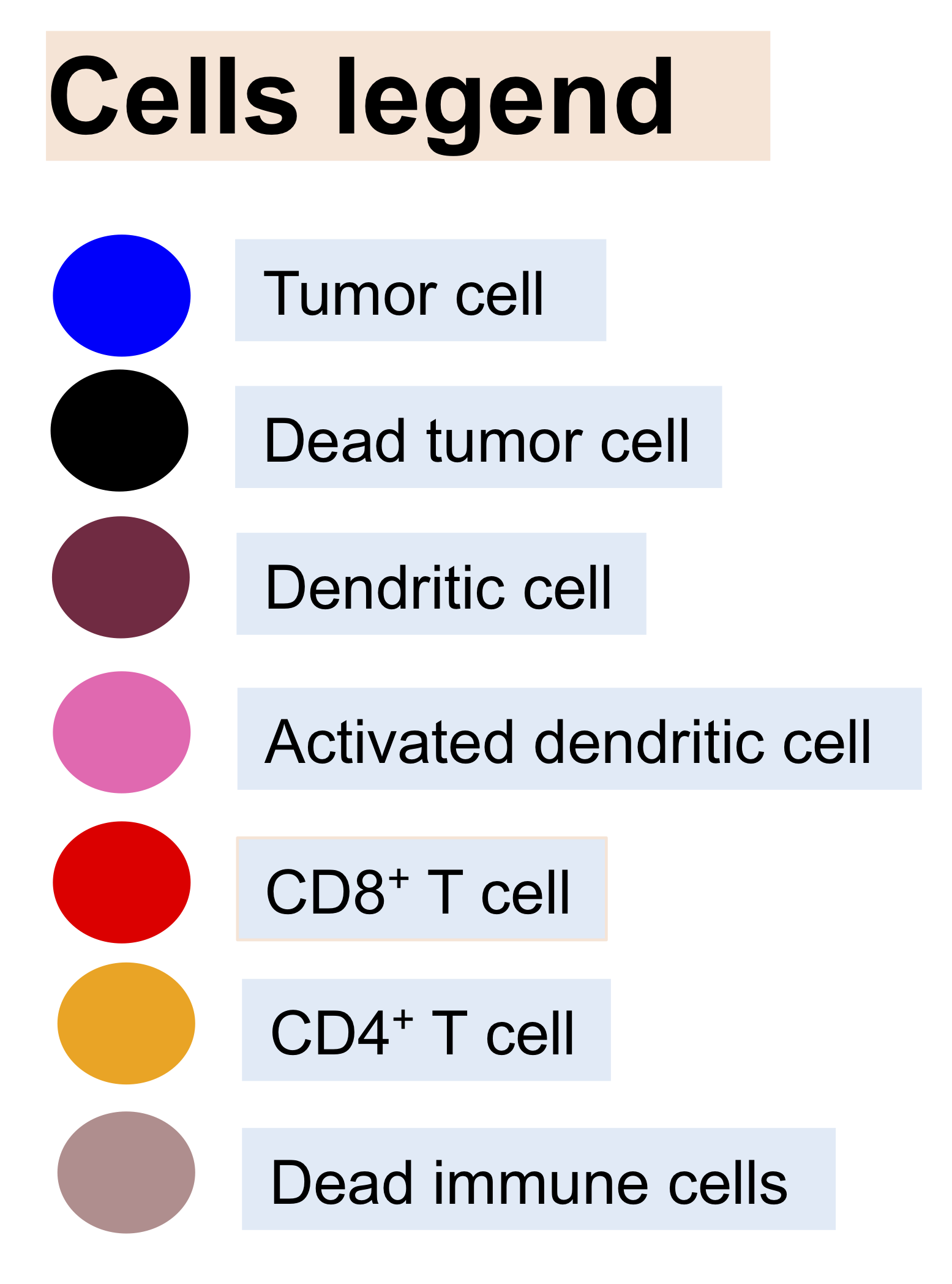
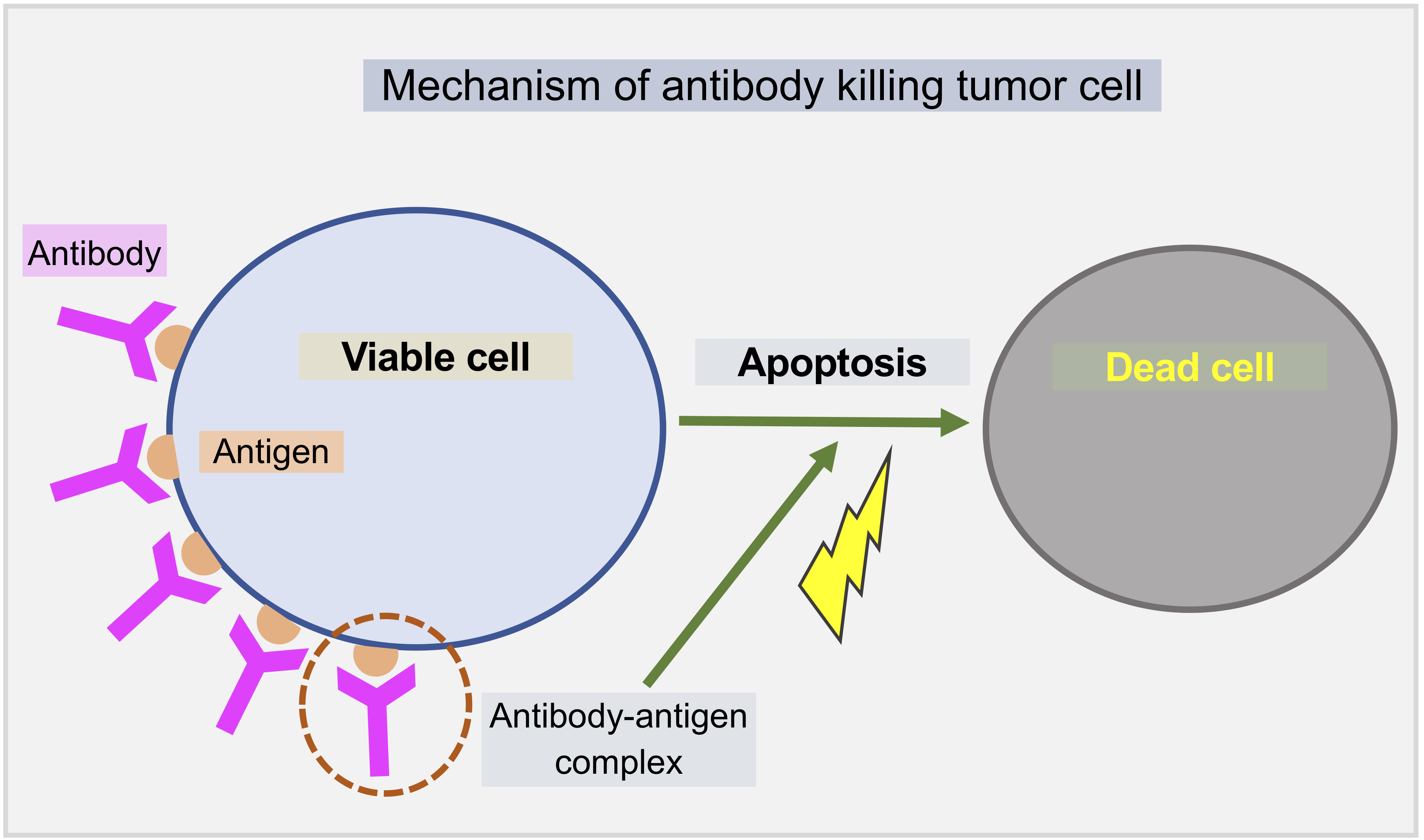
A model of mRNA vaccine-loaded lipid nanoparticles (LNPs) for cancer immunotherapy is developed by extending and adapting cancer nanobio project (https://nanohub.org/resources/pc4nanobio) and COVID19 project (https://doi.org/10.1101/2020.04.02.019075). The nano-immunotherapy model is used to perform preliminary explorations of the activation and immune response through exploring tumor cell growth, LNP-dendritic cell interactions (LNP binding dynamics, intracellular mRNA message translation dynamics), activation of immune systems (recruitment of CD8+, CD4+ T cells and antibody), and interactions between CD8+ T, antibody and tumor cells.
See below model diagrams:
Powered by
This software is powered by PhysiCell [1-2], a powerful simulation tool that combines multi-substrate diffusive transport and off-lattice cell models. PhysiCell is BSD-licensed, and available at:
- GitHub releases: https://github.com/MathCancer/PhysiCell/releases
- SourceForge downloads: https://sourceforge.net/projects/physicell/
It is a C++, cross-platform code with minimal software dependencies. It has been tested and deployed in Linux, BSD, OSX, Windows, and other environments, using the standard g++ compiler.
See http://PhysiCell.MathCancer.org.
The Jupyter-based GUI was auto-generated by xml2jupyter [3], a technique to create graphical user interfaces for command-line scientific applications.
References
[1] Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, Macklin P (2018) PhysiCell: An open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput Biol 14(2): e1005991. https://dx.doi.org/10.1371/journal.pcbi.1005991
[2] Ghaffarizadeh A, Friedman SH, Macklin P (2016) BioFVM: an efficient, parallelized diffusive transport solver for 3-D biological simulations. Bioinformatics 32(8):1256-8. https://dx.doi.org/10.1093/bioinformatics/btv730
[3] Heiland R, Mishler D, Zhang T, Bower E, Macklin P (2019) Xml2jupyter: Mapping parameters between XML and Jupyter widgets. J Open Source Software 4(39):1408. https://dx.doi.org/10.21105/joss.01408
Cite this work
Researchers should cite this work as follows: