|
Issue 48

At the close of another year, we gratefully pause to wish you a warm and happy holiday season. Thank you for being a part of the nanoHUB community. We appreciate your ongoing support and look forward to a productive new year. nanoHUB NewsNew Machine Learning Technique
Saaketh Desai and Alejandro Strachan, deputy director of nanoHUB, identified a way to use machine learning to reduce the time it takes to discover physical laws. Their study was published in Scientific Reports.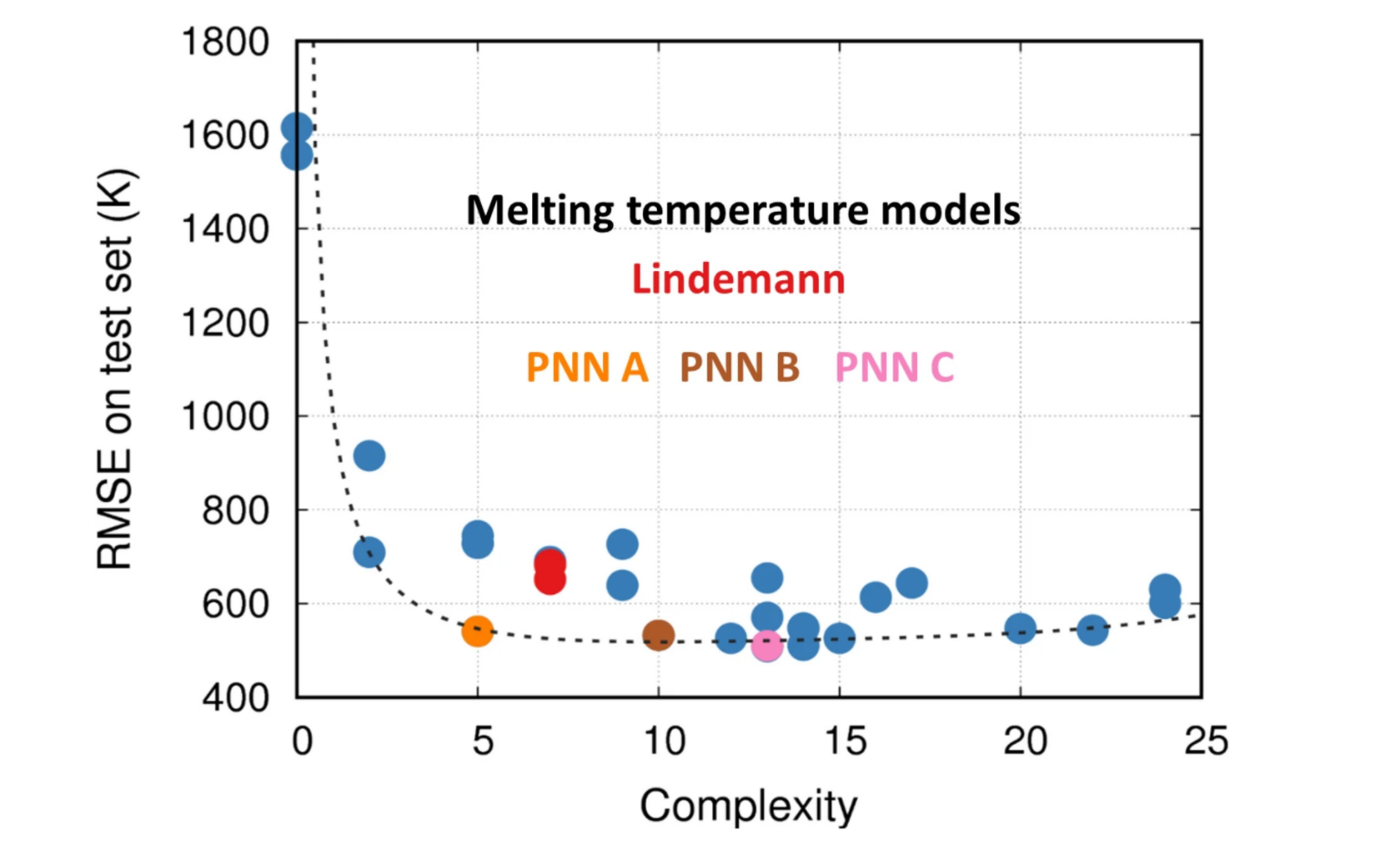
Based on their findings, they developed the Parsimonious neural networks tool, found in nanoHUB. This tool allows users to achieve simpler and more interpretable machine learning models.
Read more about the new machine learning technique here. EventsnanoBIO Simulation in Education Workshop Series
Join nanoBIO for their upcoming Spring program on simulation in education, beginning February 3, 2022. This workshop series is intended for researchers and educators to examine implementations of nanoBIO and other cloud-based applications (apps) in educational settings. Each workshop will include a presentation of the use of one or more apps along with a discussion of supporting materials such as problem sets. Time will be reserved so that workshop attendees can either offer insights for improvements or discuss how to modify the messaging for a different context.
Title: nanoBIO Simulation in Education Workshop Series
Dates: Every other Thursday at 3:00 p.m. EST, beginning February 3, 2022
Register for this series here.
For more information, please visit the nanoBIO seminar page. Recitation Series using nanoHUB Tools for Semiconductor Education
We invite all faculty and teaching assistants with an interest in learning about nanoHUB's semiconductor simulation tools to join our Recitation Series using nanoHUB Tools for Semiconductor Education. Dr. Gerhard Klimeck will introduce the ABACUS tool suite in nanoHUB. He developed these tools to teach his standard semiconductor device class at Purdue University. These tools include simulations of crystal structures and lattices, bandstructure and band models, bulk semiconductors, PN Junctions, Bipolar Junction Transistors, MOS Capacitors, and MOSFETs. The ABACUS group page highlights various features and animations and includes recordings to previous workshops.
Registration for the remaining four sessions is now open at the links below:
Recitation Series Schedule:
New Resources
Antonio Zdanis Neher created this application under the guidance of Dr. James Glazier and Josua Aponte-Serrano as part of his work in a Computational Bioengineering class. It is designed to walk users through a mathematical model representing the spread of Tuberculosis through a city population with special consideration given to the effects of vaccination, social distancing, and proper antibiotic treatment on epidemic progression. It is Antonio's hope that users of this app will come away with new knowledge of Tuberculosis, the rising threat of antibiotic resistance and generalizable lessons that can be applied to a multitude of epidemics.
|
CompuCell3D is a flexible scriptable modeling environment, which allows the rapid construction of sharable Virtual Tissue in-silico simulations of a wide variety of multi-scale, multi-cellular problems including angiogenesis, bacterial colonies, cancer, developmental biology, evolution, the immune system, tissue engineering, toxicology and even non-cellular soft materials. It was developed by Juliano Ferrari Gianlupi and Dr. T.J. Sego.
|
|
CompuCell3D models have been used to solve basic biological problems, to develop medical therapies, to assess modes of action of toxicants and to design engineered tissues. CompuCell3D makes Virtual Tissue modeling and simulation accessible to users without extensive software development or programming experience. It uses Cellular Potts Model to model cell behavior. v. 4 is now available here.
To learn more about CompuCell3D, check out their Mini-workshop on CompuCell3D Virtual-Tissue Simulations of Infection:
|
|





